Utilisateur:JeanSolPartre/Histone
Les histones sont des protéines localisées dans le noyau des cellules eucaryotes[1] et dans les archées. Elles sont les principaux constituants protéiques des chromosomes. Elles sont en effet étroitement associées à l’ADN dont elles permettent la compaction, cette action forme des structures appelées nucléosomes : l'ADN est enroulé autour des histones comme du fil autour d'une bobine[2][3]. Les histones sont très riches en acides aminés basiques (lysine et arginine), dont la charge positive à pH physiologique permet une interaction forte avec les groupements phosphate de l'ADN qui portent des charges négatives.
Histoire
[modifier | modifier le code]Les histones ont été découvertes en 1884 par Albrecht Kossel[4]. Le terme « histone » dérive de l’allemand Histon, mot à l’origine est incertaine venant peut-être du grec histanai ou histos.
Au début des années 1960, avant que l’on découvre que les histones sont fortement conservées dans le monde vivant et qu’il en existe différents types, James F. Bonner et ses collaborateurs ont lancé une étude de ces protéines
In the early 1960s, before the types of histones were known and before histones were known to be highly conserved across taxonomically diverse organisms, James F. Bonner and his collaborators began a study of these proteins that were known to be tightly associated with the DNA in the nucleus of higher organisms.[5] Bonner and his postdoctoral fellow Ru Chih C. Huang showed that isolated chromatin would not support RNA transcription in the test tube, but if the histones were extracted from the chromatin, RNA could be transcribed from the remaining DNA.[6] Their paper became a citation classic.[6] Paul T'so and James Bonner had called together a World Congress on Histone Chemistry and Biology in 1964, in which it became clear that there was no consensus on the number of kinds of histone and that no one knew how they would compare when isolated from different organisms.[7][5] Bonner and his collaborators then developed methods to separate each type of histone, purified individual histones, compared amino acid compositions in the same histone from different organisms, and compared amino acid sequences of the same histone from different organisms in collaboration with Emil Smith from UCLA.[8] For example, they found Histone IV sequence to be highly conserved between peas and calf thymus.[8] However, their work on the biochemical characteristics of individual histones did not reveal how the histones interacted with each other or with DNA to which they were tightly bound.[7]
Also in the 1960s, Vincent Allfrey and Alfred Mirsky had suggested, based on their analyses of histones, that acetylation and methylation of histones could provide a transcriptional control mechanism, but did not have available the kind of detailed analysis that later investigators were able to conduct to show how such regulation could be gene-specific.[9] Until the early 1990s, histones were dismissed by most as inert packing material for eukaryotic nuclear DNA, a view based in part on the models of Mark Ptashne and others, who believed that transcription was activated by protein-DNA and protein-protein interactions on largely naked DNA templates, as is the case in bacteria.
During the 1980s, Yahli Lorch and Roger Kornberg[10] showed that a nucleosome on a core promoter prevents the initiation of transcription in vitro, and Michael Grunstein[11] demonstrated that histones repress transcription in vivo, leading to the idea of the nucleosome as a general gene repressor. Relief from repression is believed to involve both histone modification and the action of chromatin-remodeling complexes. Vincent Allfrey and Alfred Mirsky had earlier proposed a role of histone modification in transcriptional activation,[12] regarded as a molecular manifestation of epigenetics. Michael Grunstein[13] and David Allis[14] found support for this proposal, in the importance of histone acetylation for transcription in yeast and the activity of the transcriptional activator Gcn5 as a histone acetyltransferase.
The discovery of the H5 histone appears to date back to the 1970s,[15] and it is now considered an isoform of Histone H1.[3][16][17][18]
Structure et typologie des histones
[modifier | modifier le code]
Structure des histones
[modifier | modifier le code]Les histones sont de petites protéines basiques de masse moléculaire comprise entre 13 et 15 kDa. Elles sont caractérisées par un domaine C-terminal globulaire, le domaine histone-fold. Ce domaine, très conservé depuis les archées jusqu’aux eucaryotes supérieurs, consiste en trois hélices α séparées par deux courtes boucles[19]. Il permet la dimérisation des histones selon un motif dit en poignée de main, qui sert de base à l’assemblage du nucléosome[20].
Le domaine histone-fold est retrouvé dans de nombreuses protéines autres que les histones, et définit la famille des protéines dite histone-like[21].
Les extrémités N-terminales des histones ne sont pas structurées et dépassent à l'extérieur de l'ADN dans le nucléosome assemblé. Ces extrémités sont accessibles à des enzymes de modification.
Classes d’histones
[modifier | modifier le code]Les histones comprennent cinq classes de protéines, regroupées en deux catégories en fonction de leur rôle dans la formation du nucléosome :
- les histones des classes H2A, H2B, H3 et H4 constituent les histones « de cœur », ainsi nommées car elles forment la particule de cœur du nucléosome : deux histones de chaque classe s’associent en un octamère autour duquel s’enroule le double-brin d’ADN sur environ cent cinquante paires de bases ;
- les histones de la classe H1/H5 sont les histones dites « de liaison » : une histone de cette classe lie l’ADN à l’endroit où celui-ci entre et sort de la particule de cœur, et « scelle » ainsi le nucléosome[22].
Chaque classe comprend plusieurs sous-types (exceptée la classe H4) distingués en fonction de leur profil d’expression :
- les sous-types dont l’expression est couplée à la réplication de l’ADN ;
- les sous-types dont l’expression est indépendante de la réplication de l’ADN ;
- les sous-types exprimés spécifiquement dans certains tissus ou à certaines étapes du développement.
Dans les cellules cyclantes, les sous-types dont l’expression est couplée à la réplication sont majoritaires dans la chromatine, c’est pourquoi on les qualifie d’histones conventionnelles ou canoniques. Par opposition, les autres sous-types sont qualifiés de « variants d’histones » ; ils représentent généralement moins de 10 % des histones totales dans les cellules cyclantes, mais cette proportion peut atteindre 50 % dans les cellules différenciées[23],[24].
Les gènes codant les histones conventionnelles sont généralement présents dans le génome en de multiples copies organisées en clusters — par exemple chez l’homme, trois clusters sur les chromosomes 1 et 6 ; chez la souris, trois clusters sur les chromosomes 3, 11 et 13[25]. Ils présentent des caractéristiques atypiques pour des gènes eucaryotes, comme l’absence d’introns et une terminaison de la transcription signalée par une structure de type tige-boucle au lieu d’un signal de polyadénylation. À l’opposé, les gènes codants les variants d’histones ne sont présents qu’en une ou deux copies et sont répartis isolément dans tout le génome[26]; Ils possèdent tous au moins un intron et un signal de poly-adénylation.
Variants d'histones
[modifier | modifier le code]Dans plusieurs espèces eucaryotes, des variants d’histones ont été découverts. Ces variants ont une séquence qui diffère de celle des histones conventionnelles sur quelques résidus seulement (cas des variants dits homéomorphes), ou sur des portions plus importantes de la protéine (cas des variants hétéromorphes).
Les variants d’histones jouent des rôles majeurs dans différents aspects de la biologie tels que la réparation de l’ADN[27],[28], l’organisation centromérique[29], l’inactivation du chromosome sexuel X[30], une condensation spécifique des cellules gamètes mâles[31],[32] ou le maintien de l’intégrité génomique au cours du développement[33].
Chez la plante-modèle Arabidopsis thaliana, un seul variant d’histone (H2A.Z) suffit à rendre l’individu sensible des variations de température de moins de 1°C. Cette histone modifie l’enroulement de l’ADN sur lui-même et contrôle ainsi l’accès à l’ADN de certaines molécules inhibant ou activant plusieurs dizaines de gènes. Cet effet « bio-thermostat » semble fréquent dans la nature, car également décrit chez la levure[34],[35].
Les variants d’histones de différents organismes, leur classification et leurs propriétés spécifiques sont décrits dans la base de données HistoneDB. Voici une liste des histones du génome humain :
| Type d’histone | Famille | Sous-famille | Membres |
|---|---|---|---|
| Histones de liaison | H1 | H1F | H1F0, H1FNT, H1FOO, H1FX |
| H1H1 | HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H1T | ||
| Histones « de cœur » | H2A | H2AF | H2AFB1, H2AFB2, H2AFB3, H2AFJ, H2AFV, H2AFX, H2AFY, H2AFY2, H2AFZ |
| H2A1 | HIST1H2AA, HIST1H2AB, HIST1H2AC, HIST1H2AD, HIST1H2AE, HIST1H2AG, HIST1H2AI, HIST1H2AJ, HIST1H2AK, HIST1H2AL, HIST1H2AM | ||
| H2A2 | HIST2H2AA3, HIST2H2AC | ||
| H2B | H2BF | H2BFM, H2BFS, H2BFWT | |
| H2B1 | HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO | ||
| H2B2 | HIST2H2BE | ||
| H3 | H3A1 | HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J | |
| H3A2 | HIST2H3C | ||
| H3A3 | HIST3H3 | ||
| H4 | H41 | HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L | |
| H44 | HIST4H4 |
Nucléosomes et condensation de l’ADN
[modifier | modifier le code]Il y a environ cinquante-quatre paires de bases entre deux nucléosomes, cette valeur variant selon les espèces (par exemple, on en compte cent soixante-cinq pour la levure). Le niveau suivant de compaction de l'ADN fait intervenir d'autres protéines dites « non histones. »
Le degré de condensation de l'ADN autour des nucléosomes d'histones et des protéines non histones est variable le long des chromosomes, dans la chromatine. Il est faible dans l'euchromatine que l'on dit « ouverte » et accessible à la machinerie des ARN polymérases. Il est élevé dans l'hétérochromatine, que l'on dit « fermée » et « inaccessible » à la machinerie de transcription.
Ce degré de condensation est regulé par des modifications des extrémités N-terminales des histones, comme des phosphorylations, acétylations, méthylations, ubiquitinations, sumoylations, etc. l'ensemble de ces modifications étant catalysées par des enzymes spécifiques. Les modifications covalentes des histones agiraient soit directement en modifiant la compaction de l'enroulement d'ADN autour des nucléosomes, soit indirectement en constituant des « marques » permettant le recrutement de protéines capables de modifier la structure de la chromatine. Le modèle des modifications covalentes des histones agissant comme un code (le « code des histones ») a été proposé par Strahl et Allis en 2000 dans la revue Nature[36]. Cependant, ce code est, semble-t-il, loin d'être universel et plutôt relativement spécifique selon les gènes et les cellules considérés.
Histones act as spools around which DNA winds. This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nuclei: the compacted molecule is 40,000 times shorter than an unpacked molecule.
Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to 1 in human DNA). For example, each human diploid cell (containing 23 pairs of chromosomes) has about 1.8 meters of DNA; wound on the histones, the diploid cell has about 90 micrometers (0.09 mm) of chromatin. When the diploid cells are duplicated and condensed during mitosis, the result is about 120 micrometers of chromosomes.[27]
Dans les spermatozoïdes, es histones sont remplacées par des protamines, protéines riches en arginine et en cystéine.
La richesse en cystéine permet la formation de pont disulfure. Cette structure protège l'ADN lors d'éventuels déplacements liés à la fécondation.
Structure des nucléosomes
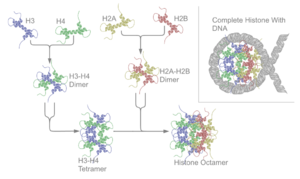
[modifier | modifier le code]The resulting four distinct dimers then come together to form one octameric nucleosome core, approximately 63 Angstroms in diameter (a solenoid (DNA)-like particle). Around 146 base pairs (bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across.[37] The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place[38] and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA separating each pair of nucleosomes (also referred to as linker DNA). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer, forming two nearly symmetrical halves by tertiary structure (C2 symmetry; one macromolecule is the mirror image of the other).[37] The H2A-H2B dimers and H3-H4 tetramer also show pseudodyad symmetry. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution, all featuring a 'helix turn helix turn helix' motif (DNA-binding protein motif that recognize specific DNA sequence). They also share the feature of long 'tails' on one end of the amino acid structure - this being the location of post-translational modification (see below).[39]
Archaeal histone only contains a H3-H4 like dimeric structure made out of the same protein. Such dimeric structures can stack into a tall superhelix ("supernucleosome") onto which DNA coils in a manner similar to nucleosome spools.[40] Only some archaeal histones have tails.[41]
It has been proposed that histone proteins are evolutionarily related to the helical part of the extended AAA+ ATPase domain, the C-domain, and to the N-terminal substrate recognition domain of Clp/Hsp100 proteins. Despite the differences in their topology, these three folds share a homologous helix-strand-helix (HSH) motif.[39]
Using an electron paramagnetic resonance spin-labeling technique, British researchers measured the distances between the spools around which eukaryotic cells wind their DNA. They determined the spacings range from 59 to 70 Å.[42]
In all, histones make five types of interactions with DNA:
- Helix-dipoles form alpha-helixes in H2B, H3, and H4 cause a net positive charge to accumulate at the point of interaction with negatively charged phosphate groups on DNA
- Hydrogen bonds between the DNA backbone and the amide group on the main chain of histone proteins
- Nonpolar interactions between the histone and deoxyribose sugars on DNA
- Salt bridges and hydrogen bonds between side chains of basic amino acids (especially lysine and arginine) and phosphate oxygens on DNA
- Non-specific minor groove insertions of the H3 and H2B N-terminal tails into two minor grooves each on the DNA molecule
The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to their water solubility.
Histones are subject to post translational modification by enzymes primarily on their N-terminal tails, but also in their globular domains.[43][44] Such modifications include methylation, citrullination, acetylation, phosphorylation, SUMOylation, ubiquitination, and ADP-ribosylation. This affects their function of gene regulation.
In general, genes that are active have less bound histone, while inactive genes are highly associated with histones during interphase.[45] It also appears that the structure of histones has been evolutionarily conserved, as any deleterious mutations would be severely maladaptive. All histones have a highly positively charged N-terminus with many lysine and arginine residues.
Modifications post-traductionnelles des histones
[modifier | modifier le code]Le code des histones établit un lien direct entre la modification de certains résidus de la queue des histones qui crée des liaisons pour des effecteurs protéiques et l’état transcriptionnel de la chromatines[1].
Une modification d’histone peut en influencer une autre de manière synergique ou antagoniste ; c’est un mécanisme qui génère et stabilise des empreintes spécifiques.
L’acétylation (ajout de groupement acétyl) s’effectue sur certains résidus lysine précis par des enzymes nommées histones acétyl transférases (HAT).
Elle diminue généralement l’interaction inter-nucléosomes et entre les queues des histones et le fragment d’ADN qui fait le lien entre les nucléosomes. Ceci entraîne le relâchement de la chromatine, la faisant passer à l'état euchromatinien, et permet ainsi une meilleure accessibilité aux autres facteurs. L’acétylation est associée à une activation de la transcription et est facilement réversible grâce à l’action des histones désacétylases (HDAC).
La méthylation, quant à elle, peut s’effectuer soit sur des lysines soit sur des arginines et peut consister en l'ajout d'un, de deux ou de trois groupements méthyls.
Selon les résidus méthylés et le nombre de groupement ajouté elle est associée à une activation ou une répression de la transcription. Longtemps considérée comme statique, la méthylation des histones s'avère être une modification réversible impliquée dans un processus dynamique, bien que plus stable que l'acétylation et la phosphorylation. Un nombre croissant d'histones déméthylases sont identifiés[46].
De manière générale, ces deux types de modifications sont antagonistes, et la désacétylation des lysines doit précéder leur méthylation. Cet antagonisme entraine la mise en place d'un certain équilibre dynamique entre les territoires hétérochromatiniens (généralement non exprimables et méthylés sur certains acides aminés clés) et euchromatiniens (généralement exprimables et acétylés). Par exemple, la Lysine 9 de l'histone H3 est connue pour être associée à une répression de la chromatine environnante lorsqu'elle est méthylée. Cette méthylation est reconnue par une protéine, HP1, qui se fixe donc sur H3 méthylée. À son tour, HP1 attire la protéine Suv39, une Histone MethylTransférase, qui pourra méthyler la lysine 9 de l'histone H3 du nucléosome voisin, et ainsi de suite. On voit donc, comment, de proche en proche, les histones H3 seront méthylées et la chromatine sera condensée. Cependant, cette invasion hétérochromatinienne sera stoppée si la lysine 9 de H3 rencontrée est déjà acétylée. Ainsi se met en place un équilibre compétitif entre domaines chromatiniens exprimés et réprimés.
Les modifications des queues d'histones jouent le rôle de « marques » épigénétiques qui entraînent le recrutement de différentes classes de protéines, puisque les lysines acétylées ou méthylées sont reconnues par des domaines protéiques différents. De plus, le recrutement de certains facteurs au niveau de la chromatine nécessite l’existence préalable de modifications d’histones et de protéines déjà liées. Le code des histones est donc interprété dans le contexte d’autres facteurs associés à la chromatine et c’est la combinaison d’interaction entre les histones modifiées et d’autres facteurs qui détermine si une protéine est recrutée à la chromatine
Histones undergo posttranslational modifications that alter their interaction with DNA and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome, which can be covalently modified at several places. Modifications of the tail include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, citrullination, and ADP-ribosylation. The core of the histones H2A and H2B can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code".[36][47] Histone modifications act in diverse biological processes such as gene regulation, DNA repair, chromosome condensation (mitosis) and spermatogenesis (meiosis).[48]
The common nomenclature of histone modifications is:
- The name of the histone (e.g., H3)
- The single-letter amino acid abbreviation (e.g., K for Lysine) and the amino acid position in the protein
- The type of modification (Me: methyl, P: phosphate, Ac: acetyl, Ub: ubiquitin)
- The number of modifications (only Me is known to occur in more than one copy per residue. 1, 2 or 3 is mono-, di- or tri-methylation)
So H3K4me1 denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal) of the H3 protein.
| Type of modification |
Histone | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| H3K4 | H3K9 | H3K14 | H3K27 | H3K79 | H3K36 | H4K20 | H2BK5 | H2BK20 | |
| mono-methylation | activation[49] | activation[50] | activation[50] | activation[50][51] | activation[50] | activation[50] | |||
| di-methylation | repression[52] | repression[52] | activation[51] | ||||||
| tri-methylation | activation[53] | repression[50] | repression[50] | activation,[51] repression[50] |
activation | repression[52] | |||
| acetylation | activation[54] | activation[53] | activation[53] | activation[55] | activation | ||||
A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a histone code, whereby combinations of histone modifications have specific meanings. However, most functional data concerns individual prominent histone modifications that are biochemically amenable to detailed study.

Chemistry of histone modifications
[modifier | modifier le code]Lysine methylation
[modifier | modifier le code]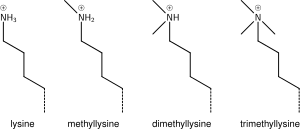
The addition of one, two, or many methyl groups to lysine has little effect on the chemistry of the histone; methylation leaves the charge of the lysine intact and adds a minimal number of atoms so steric interactions are mostly unaffected. However, proteins containing Tudor, chromo or PHD domains, amongst others, can recognise lysine methylation with exquisite sensitivity and differentiate mono, di and tri-methyl lysine, to the extent that, for some lysines (e.g.: H4K20) mono, di and tri-methylation appear to have different meanings. Because of this, lysine methylation tends to be a very informative mark and dominates the known histone modification functions.
Glutamine serotonylation
[modifier | modifier le code]Recently it has been shown, that the addition of a serotonin group to the position 5 glutamine of H3, happens in serotonergic cells such as neurons. This is part of the differentiation of the serotonergic cells. This post-translational modification happens in conjunction with the H3K4me3 modification. The serotonylation potentiates the binding of the general transcription factor TFIID to the TATA box.[56]
Arginine methylation
[modifier | modifier le code]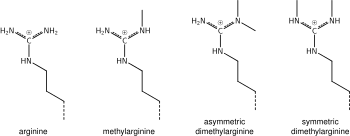
What was said above of the chemistry of lysine methylation also applies to arginine methylation, and some protein domains—e.g., Tudor domains—can be specific for methyl arginine instead of methyl lysine. Arginine is known to be mono- or di-methylated, and methylation can be symmetric or asymmetric, potentially with different meanings.
Arginine citrullination
[modifier | modifier le code]Enzymes called peptidylarginine deiminases (PADs) hydrolyze the imine group of arginines and attach a keto group, so that there is one less positive charge on the amino acid residue. This process has been involved in the activation of gene expression by making the modified histones less tightly bound to DNA and thus making the chromatin more accessible.[57] PADs can also produce the opposite effect by removing or inhibiting mono-methylation of arginine residues on histones and thus antagonizing the positive effect arginine methylation has on transcriptional activity.[58]
Lysine acetylation
[modifier | modifier le code]
Addition of an acetyl group has a major chemical effect on lysine as it neutralises the positive charge. This reduces electrostatic attraction between the histone and the negatively charged DNA backbone, loosening the chromatin structure; highly acetylated histones form more accessible chromatin and tend to be associated with active transcription. Lysine acetylation appears to be less precise in meaning than methylation, in that histone acetyltransferases tend to act on more than one lysine; presumably this reflects the need to alter multiple lysines to have a significant effect on chromatin structure. The modification includes H3K27ac.
Serine/threonine/tyrosine phosphorylation
[modifier | modifier le code]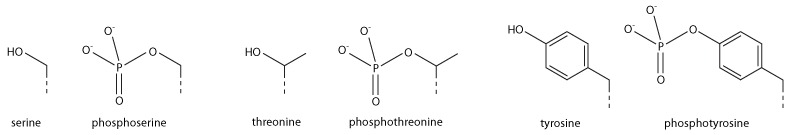
Addition of a negatively charged phosphate group can lead to major changes in protein structure, leading to the well-characterised role of phosphorylation in controlling protein function. It is not clear what structural implications histone phosphorylation has, but histone phosphorylation has clear functions as a post-translational modification, and binding domains such as BRCT have been characterised.
Fonctions des modifications post-traductionnelles : le code des histones
[modifier | modifier le code]Functions in transcription
[modifier | modifier le code]Most well-studied histone modifications are involved in control of transcription.
Actively transcribed genes
[modifier | modifier le code]Two histone modifications are particularly associated with active transcription:
- Trimethylation of H3 lysine 4 (H3K4me3)
- This trimethylation occurs at the promoter of active genes[59][60][61] and is performed by the COMPASS complex.[62][63][64] Despite the conservation of this complex and histone modification from yeast to mammals, it is not entirely clear what role this modification plays. However, it is an excellent mark of active promoters and the level of this histone modification at a gene's promoter is broadly correlated with transcriptional activity of the gene. The formation of this mark is tied to transcription in a rather convoluted manner: early in transcription of a gene, RNA polymerase II undergoes a switch from initiating’ to ‘elongating’, marked by a change in the phosphorylation states of the RNA polymerase II C terminal domain (CTD). The same enzyme that phosphorylates the CTD also phosphorylates the Rad6 complex,[65][66] which in turn adds a ubiquitin mark to H2B K123 (K120 in mammals).[67] H2BK123Ub occurs throughout transcribed regions, but this mark is required for COMPASS to trimethylate H3K4 at promoters.[68][69]
- Trimethylation of H3 lysine 36 (H3K36me3)
- This trimethylation occurs in the body of active genes and is deposited by the methyltransferase Set2.[70] This protein associates with elongating RNA polymerase II, and H3K36Me3 is indicative of actively transcribed genes.[71] H3K36Me3 is recognised by the Rpd3 histone deacetylase complex, which removes acetyl modifications from surrounding histones, increasing chromatin compaction and repressing spurious transcription.[72][73][74] Increased chromatin compaction prevents transcription factors from accessing DNA, and reduces the likelihood of new transcription events being initiated within the body of the gene. This process therefore helps ensure that transcription is not interrupted.
Repressed genes
[modifier | modifier le code]Three histone modifications are particularly associated with repressed genes:
- Trimethylation of H3 lysine 27 (H3K27me3)
- This histone modification is depositied by the polycomb complex PRC2.[75] It is a clear marker of gene repression,[76] and is likely bound by other proteins to exert a repressive function. Another polycomb complex, PRC1, can bind H3K27me3[76] and adds the histone modification H2AK119Ub which aids chromatin compaction.[77][78] Based on this data it appears that PRC1 is recruited through the action of PRC2, however, recent studies show that PRC1 is recruited to the same sites in the absence of PRC2.[79][80]
- Di and tri-methylation of H3 lysine 9 (H3K9me2/3)
- H3K9me2/3 is a well-characterised marker for heterochromatin, and is therefore strongly associated with gene repression. The formation of heterochromatin has been best studied in the yeast Schizosaccharomyces pombe, where it is initiated by recruitment of the RNA-induced transcriptional silencing (RITS) complex to double stranded RNAs produced from centromeric repeats.[81] RITS recruits the Clr4 histone methyltransferase which deposits H3K9me2/3.[82] This process is called histone methylation. H3K9Me2/3 serves as a binding site for the recruitment of Swi6 (heterochromatin protein 1 or HP1, another classic heterochromatin marker)[83][84] which in turn recruits further repressive activities including histone modifiers such as histone deacetylases and histone methyltransferases.[85]
- Trimethylation of H4 lysine 20 (H4K20me3)
- This modification is tightly associated with heterochromatin,[86][87] although its functional importance remains unclear. This mark is placed by the Suv4-20h methyltransferase, which is at least in part recruited by heterochromatin protein 1.[86]
Bivalent promoters
[modifier | modifier le code]Analysis of histone modifications in embryonic stem cells (and other stem cells) revealed many gene promoters carrying both H3K4Me3 and H3K27Me3, in other words these promoters display both activating and repressing marks simultaneously. This peculiar combination of modifications marks genes that are poised for transcription; they are not required in stem cells, but are rapidly required after differentiation into some lineages. Once the cell starts to differentiate, these bivalent promoters are resolved to either active or repressive states depending on the chosen lineage.[88]
Other functions
[modifier | modifier le code]DNA damage
[modifier | modifier le code]Marking sites of DNA damage is an important function for histone modifications. It also protects DNA from getting destroyed by ultraviolet radiation of sun.
- Phosphorylation of H2AX at serine 139 (γH2AX)
- Phosphorylated H2AX (also known as gamma H2AX) is a marker for DNA double strand breaks,[89] and forms part of the response to DNA damage.[90][91] H2AX is phosphorylated early after detection of DNA double strand break, and forms a domain extending many kilobases either side of the damage.[89][92][93] Gamma H2AX acts as a binding site for the protein MDC1, which in turn recruits key DNA repair proteins[94] (this complex topic is well reviewed in[95]) and as such, gamma H2AX forms a vital part of the machinery that ensures genome stability.
- Acetylation of H3 lysine 56 (H3K56Ac)
- H3K56Acx is required for genome stability.[96][97] H3K56 is acetylated by the p300/Rtt109 complex,[98][99][100] but is rapidly deacetylated around sites of DNA damage. H3K56 acetylation is also required to stabilise stalled replication forks, preventing dangerous replication fork collapses.[101][102] Although in general mammals make far greater use of histone modifications than microorganisms, a major role of H3K56Ac in DNA replication exists only in fungi, and this has become a target for antibiotic development.[103]
DNA repair
[modifier | modifier le code]- Trimethylation of H3 lysine 36 (H3K36me3)
H3K36me3 has the ability to recruit the MSH2-MSH6 (hMutSα) complex of the DNA mismatch repair pathway.[104] Consistently, regions of the human genome with high levels of H3K36me3 accumulate less somatic mutations due to mismatch repair activity.[105]
Chromosome condensation
[modifier | modifier le code]- Phosphorylation of H3 at serine 10 (phospho-H3S10)
- The mitotic kinase aurora B phosphorylates histone H3 at serine 10, triggering a cascade of changes that mediate mitotic chromosome condensation.[106][107] Condensed chromosomes therefore stain very strongly for this mark, but H3S10 phosphorylation is also present at certain chromosome sites outside mitosis, for example in pericentric heterochromatin of cells during G2. H3S10 phosphorylation has also been linked to DNA damage caused by R-loop formation at highly transcribed sites.[108]
- Phosphorylation H2B at serine 10/14 (phospho-H2BS10/14)
- Phosphorylation of H2B at serine 10 (yeast) or serine 14 (mammals) is also linked to chromatin condensation, but for the very different purpose of mediating chromosome condensation during apoptosis.[109][110] This mark is not simply a late acting bystander in apoptosis as yeast carrying mutations of this residue are resistant to hydrogen peroxide-induced apoptotic cell death.
Addiction
[modifier | modifier le code]Epigenetic modifications of histone tails in specific regions of the brain are of central importance in addictions.[111][112][113] Once particular epigenetic alterations occur, they appear to be long lasting "molecular scars" that may account for the persistence of addictions.[111]
Cigarette smokers (about 15% of the US population) are usually addicted to nicotine.[114] After 7 days of nicotine treatment of mice, acetylation of both histone H3 and histone H4 was increased at the FosB promoter in the nucleus accumbens of the brain, causing 61% increase in FosB expression.[115] This would also increase expression of the splice variant Delta FosB. In the nucleus accumbens of the brain, Delta FosB functions as a "sustained molecular switch" and "master control protein" in the development of an addiction.[116][117]
About 7% of the US population is addicted to alcohol. In rats exposed to alcohol for up to 5 days, there was an increase in histone 3 lysine 9 acetylation in the pronociceptin promoter in the brain amygdala complex. This acetylation is an activating mark for pronociceptin. The nociceptin/nociceptin opioid receptor system is involved in the reinforcing or conditioning effects of alcohol.[118]
Methamphetamine addiction occurs in about 0.2% of the US population.[119] Chronic methamphetamine use causes methylation of the lysine in position 4 of histone 3 located at the promoters of the c-fos and the C-C chemokine receptor 2 (ccr2) genes, activating those genes in the nucleus accumbens (NAc).[120] c-fos is well known to be important in addiction.[121] The ccr2 gene is also important in addiction, since mutational inactivation of this gene impairs addiction.[120]
Conservation across species
[modifier | modifier le code]Histones are believed to have evolved from ribosomal proteins with which they share much in common, both being short and basic proteins. [122]
Histones are found in the nuclei of eukaryotic cells, and in certain Archaea, namely Proteoarchaea and Euryarchaea, but not in bacteria.[41] The unicellular algae known as dinoflagellates were previously thought to be the only eukaryotes that completely lack histones,[123] however, later studies showed that their DNA still encodes histone genes.[124] Unlike the core histones, lysine-rich linker histone (H1) proteins are found in bacteria, otherwise known as nucleoprotein HC1/HC2.[125]
Archaeal histones may well resemble the evolutionary precursors to eukaryotic histones.[41] Histone proteins are among the most highly conserved proteins in eukaryotes, emphasizing their important role in the biology of the nucleus.[3]:939 In contrast mature sperm cells largely use protamines to package their genomic DNA, most likely because this allows them to achieve an even higher packaging ratio.[126]
There are some variant forms in some of the major classes. They share amino acid sequence homology and core structural similarity to a specific class of major histones but also have their own feature that is distinct from the major histones. These minor histones usually carry out specific functions of the chromatin metabolism. For example, histone H3-like CENPA is associated with only the centromere region of the chromosome. Histone H2A variant H2A.Z is associated with the promoters of actively transcribed genes and also involved in the prevention of the spread of silent heterochromatin.[127] Furthermore, H2A.Z has roles in chromatin for genome stability.[28] Another H2A variant H2A.X is phosphorylated at S139 in regions around double-strand breaks and marks the region undergoing DNA repair.[90] Histone H3.3 is associated with the body of actively transcribed genes.[128]
Histone synthesis
[modifier | modifier le code]The first step of chromatin structure duplication is the synthesis of histone proteins: H1, H2A, H2B, H3, H4. These proteins are synthesized during S phase of the cell cycle. There are different mechanisms which contribute to the increase of histone synthesis.
Yeast
[modifier | modifier le code]Yeast carry one or two copies of each histone gene, which are not clustered but rather scattered throughout chromosomes. Histone gene transcription is controlled by multiple gene regulatory proteins such as transcription factors which bind to histone promoter regions. In budding yeast, the candidate gene for activation of histone gene expression is SBF. SBF is a transcription factor that is activated in late G1 phase, when it dissociates from its repressor Whi5. This occurs when Whi5 is phosphorylated by Cdc8 which is a G1/S Cdk.[129] Suppression of histone gene expression outside of S phases is dependent on Hir proteins which form inactive chromatin structure at the locus of histone genes, causing transcriptional activators to be blocked.[130][131]
Metazoans
[modifier | modifier le code]In metazoans the increase in the rate of histone synthesis is due to the increase in processing of pre-mRNA to its mature form as well as decrease in mRNA degradation; this results in an increase of active mRNA for translation of histone proteins. The mechanism for mRNA activation has been found to be the removal of a segment of the 3’ end of the mRNA strand, and is dependent on association with stem-loop binding protein (SLBP).[132] SLBP also stabilizes histone mRNAs during S phase by blocking degradation by the 3’hExo nuclease.[133] SLBP levels are controlled by cell-cycle proteins, causing SLBP to accumulate as cells enter S phase and degrade as cells leave S phase. SLBP are marked for degradation by phosphorylation at two threonine residues by cyclin dependent kinases, possibly cyclin A/ cdk2, at the end of S phase.[134] Metazoans also have multiple copies of histone genes clustered on chromosomes which are localized in structures called Cajal bodies as determined by genome-wide chromosome conformation capture analysis (4C-Seq).[135]
Link between cell-cycle control machinery and histone synthesis
[modifier | modifier le code]Nuclear protein Ataxia-Telangiectasia (NPAT), also known as nuclear protein coactivator of histone transcription, is a transcription factor which activates histone gene transcription on chromosomes 1 and 6 of human cells. NPAT is also a substrate of cyclin E-Cdk2, which is required for the transition between G1 phase and S phase. NPAT activates histone gene expression only after it has been phosphorylated by the G1/S-Cdk cyclin E-Cdk2 in early S phase.[136] This shows an important regulatory link between cell-cycle control and histone synthesis.
See also
[modifier | modifier le code]
References
[modifier | modifier le code]- Annabelle Gérard, Sophie Polo et Geneviève Almouzni, « Nom de code : histones », Pour la science, no 46, (ISSN 0153-4092, lire en ligne)
- (en) Robert M. Youngson, Collins Dictionary of Human Biology, Glasgow, Écosse, Royaume-Uni, Collins, , 1re éd., 522 p. (ISBN 978-0-00-722134-9)
- (en) David R. Nelson, Michael Cox et Albert L. Lehninger, Principles of Biochemistry, San Francisco, Californie, États-Unis, W. H. Freeman, , 4e éd., 1119 p. (ISBN 978-0-7167-4339-2)
- (en) J. Murray Luck, « Histone Chemistry: the Pioneers », dans James F. Bonner et Paul O. P. T’so (dir.), The Nucleohistones, San Francisco, Californie, États-Unis, Holden-Day, , XVII-398 p., p. 3-14
- James Bonner, « Chapters from my life », Annual Review of Plant Physiology and Plant Molecular Biology, vol. 45, , p. 1-23 (DOI 10.1146/annurev.pp.45.060194.000245)
- (en) Ru-chih Huang et James Bonner, « Histone, a suppressor of chromosomal RNA synthesis. », Proceedings of the National Academy of Sciences of the United States of America, vol. 48, , p. 1216-22 (PMID 14036409, PMCID PMC220935, DOI 10.1073/pnas.48.7.1216)
- (en) James F. Bonner et Paul O. P. T’so, The Nucleohistones, San Francisco, Californie, États-Unis, Holden-Day, , XVII-398 p.
- (en) R. J. DeLange, D. M. Fambrough, E. L. Smith et J. Bonner, « Calf and pea histone IV. 3. Complete amino acid sequence of pea seedling histone IV; comparison with the homologous calf thymus histone. », Journal of Biological Chemistry, vol. 244, no 20, , p. 5669-79 (PMID 5388597)
- (en) V. G. Allfrey, R. Faulkner et A. E. Mirsky, « Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. », Proceedings of the National Academy of Sciences of the United States of America, vol. 51, , p. 786-94 (PMID 14172992, PMCID PMC300163, DOI 10.1073/pnas.51.5.786)
- (en) Y. Lorch, J. W. LaPointe et R. D. Kornberg, « Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. », Cell, vol. 49, no 2, , p. 203-10 (PMID 3568125, DOI 10.1016/0092-8674(87)90561-7)
- (en) Paul S. Kayne, Ung-Jin Kim, Min Han, Janet R. Mullen, Fuminori Yoshizaki et Michael Grunstein, « Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. », Cell, vol. 55, no 1, , p. 27-39 (PMID 3048701, DOI 10.1016/0092-8674(88)90006-2)
- (en) B. G. Pogo, V. G. Allfrey et A. E. Mirsky, « RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. », Proceedings of the National Academy of Sciences of the United States of America, vol. 55, no 4, , p. 805-12 (PMID 5219687, PMCID PMC224233, DOI 10.1073/pnas.55.4.805)
- (en) Linda K. Durrin, Randall K. Mann, Paul S. Kayne et Michael Grunstein, « Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. », Cell, vol. 65, no 6, , p. 1023-31 (PMID 2044150, DOI 10.1016/0092-8674(91)90554-c)
- (en) James E. Brownell, Jianxin Zhou, Tamara Ranalli, Ryuji Kobayashi, Diane G. Edmondson, Sharon Y. Roth et C. David Allis, « Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. », Cell, vol. 84, no 6, , p. 843-51 (PMID 8601308, DOI 10.1016/s0092-8674(00)81063-6)
- (en) Francisco J. Aviles, George E. Chapman, Geoff G. Kneale, Colyn Crane-Robinson et E. Morton Bradbury, « The conformation of histone H5. Isolation and characterisation of the globular segment. », European Journal of Biochemistry, vol. 88, no 2, , p. 363-71 (PMID 689022, DOI 10.1111/j.1432-1033.1978.tb12457.x)
- (en) National Center for Biotechnology Information, « Histone Variants Database 2.0 », sur ncbi.nlm.nih.gov (consulté le )
- (en) Manoj Bhasin, Ellis L. Reinherz et Pedro A. Reche, « Recognition and classification of histones using support vector machine. », Journal of Computational Biology, vol. 13, no 1, , p. 102-12 (PMID 16472024, DOI 10.1089/cmb.2006.13.102)
- (en) Daniel L. Hartl, David Freifelder et Leon A. Snyder, Basic Genetics, Boston, Massachusetts, États-Unis, Jones and Bartlett Publishers, , XVI-505 p. (ISBN 978-0-86720-090-4)
- (en) Gina Arents et Evangelos N. Moudrianakis, « The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization », Proceedings of the National Academy of Sciences of the United States of America, vol. 92, no 24, , p. 11170-4 (PMID 7479959, PMCID PMC40593, DOI 10.1073/pnas.92.24.11170)
- (en) Leonardo Marino-Ramirez, Maricel G. Kann, Benjamin A. Shoemaker et David Landsman, « Histone structure and nucleosome stability. », Expert Review of Proteomics, vol. 2, no 5, , p. 719-29 (PMID 16209651, PMCID PMC1831843, DOI 10.1586/14789450.2.5.719)
- (en) Steven Sullivan, Daniel Sink, Kenneth Trout, Izabela Makalowska, Patrick Taylor, Andreas Baxevanis et David Landsman, « The Histone Database », Nucleic Acids Research, vol. 30, no 1, , p. 341-2 (PMID 11752331, PMCID PMC99096, DOI 10.1093/nar/30.1.341)
- (en) Li Fan et Victoria Roberts, « Complex of linker histone H5 with the nucleosome and its implications for chromatin packing », Proceedings of the National Academy of Sciences of the United States of America, vol. 103, no 22, , p. 8384-9 (PMID 16717183, PMCID PMC1482502, DOI 10.1073/pnas.0508951103)
- (en) David Brush, Jerry B. Dodgson, Ok-Ryun Choi, Priscilla Stevens et James Douglas Engel, « Replacement variant histone genes contain intervening sequences », Molecular and Cellular Biology, vol. 5, no 6, , p. 1307-17 (PMID 2863747, PMCID PMC366859, DOI 10.1128/mcb.5.6.1307)
- (en) Dan Wells et Larry Kedes, « Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs », Proceedings of the National Academy of Sciences of the United States of America, vol. 82, no 9, , p. 2834-8 (PMID 2859593, DOI 10.1073/pnas.82.9.2834)
- (en) William Marzluff, Preetam Gongidi, Keith Woods, Jianping Jin et Lois Maltais, « The human and mouse replication-dependent histone genes », Genomics, vol. 80, no 5, , p. 487-498 (PMID 12408966, DOI 10.1006/geno.2002.6850)
- (en) Rohinton Kamakaka et Sue Biggins, « Histone variants: deviants? », Genes & Development, vol. 19, no 3, , p. 295-310 (PMID 15687254, DOI 10.1101/gad.1272805)
- (en) Christophe Redon, Duane Pilch, Emmy Rogakou, Olga Sedelnikova, Kenneth Newrock et William Bonner, « Histone H2A variants H2AX and H2AZ. », Current Opinion in Genetics & Development, vol. 12, no 2, , p. 162-9 (PMID 11893489, DOI 10.1016/s0959-437x(02)00282-4)
- (en) Pierre Billon et Jacques Côté, « Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. », Biochimica et Biophysica Acta, vol. 1819, nos 3-4, , p. 290-302 (PMID 24459731, DOI 10.1016/j.bbagrm.2011.10.004)
- (en) Daniel Foltz, Lars Jansen, Aaron Bailey, John Yates, Emily Bassett, Stacey Wood, Ben Black et Don Cleveland, « Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP », Cell, vol. 137, no 3, , p. 472-84 (PMID 19410544, PMCID PMC2747366, DOI 10.1016/j.cell.2009.02.039)
- (en) Oscar Fernandez-Capetillo, Shantha K. Mahadevaiah, Arkady Celeste, Peter J. Romanienko, R. Daniel Camerini-Otero, William M. Bonner, Katia Manova, Paul Burgoyne et André Nussenzweig, « H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis », Developmental Cell, vol. 4, no 4, , p. 497-508 (PMID 12689589, DOI 10.1016/s1534-5807(03)00093-5)
- (en) Takashi Okada, Makoto Endo, Mohan Singh et Prem Bhalla, « Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3 », Plant Journal, vol. 44, no 4, , p. 557-68 (PMID 16262706, DOI 10.1111/j.1365-313X.2005.02554.x)
- (en) Jérôme Govin, Cécile Caron, Cécile Lestrat, Sophie Rousseaux et Saadi Khochbin, « The role of histones in chromatin remodelling during mammalian spermiogenesis », European Journal of Biochemistry, vol. 271, no 17, , p. 3459-69 (PMID 15317581, DOI 10.1111/j.1432-1033.2004.04266.x)
- (en) Chuan-Wei Jang, Yoichiro Shibata, Joshua Starmer, Della Yee et Terry Magnuson, « Histone H3.3 maintains genome integrity during mammalian development. », Genes & Development, vol. 29, no 13, , p. 1377-92 (PMID 26159997, PMCID PMC4511213, DOI 10.1101/gad.264150.115)
- (en) S. Vinod Kumar et Philip A. Wigge, « H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis », Cell, vol. 140, no 1, , p. 136-47 (PMID 20079334, DOI 10.1016/j.cell.2009.11.006)
- Jean-Jacques Perrier, « Une protéine thermomètre chez les plantes », Pour la science, (lire en ligne)
- (en) Brian D. Strahl et C. David Allis, « The language of covalent histone modifications. », Nature, vol. 403, no 6765, , p. 41-5 (PMID 10638745, DOI 10.1038/47412)
- (en) Karolin Luger, Armin W. Mader, Robin K. Richmond, David F. Sargent et Timothy J. Richmond, « Crystal structure of the nucleosome core particle at 2.8 Å resolution. », Nature, vol. 389, no 6648, , p. 251-60 (PMID 9305837, DOI 10.1038/38444)
- (en) Daniel Farkas, DNA simplified : The hitchhiker’s guide to DNA, Washington, District de Columbia, États-Unis, American Association for Clinical Chemistry Press, , VII-110 p. (ISBN 978-0-915274-84-0)
- (en) Vikram Alva, Moritz Ammelburg, Johannes Soding et Andrei N. Lupas, « On the origin of the histone fold. », BMC Structural Biology, vol. 7, , p. 17 (PMID 17391511, PMCID PMC1847821, DOI 10.1186/1472-6807-7-17)
- (en) Francesca Mattiroli, Sudipta Bhattacharyya, Pamela N. Dyer, Alison E. White, Kathleen Sandman, Brett W. Burkhart, Kyle R. Byrne, Thomas Lee, Natalie G. Ahn, Thomas J. Santangelo, John N. Reeve et Karolin Luger, « Structure of histone-based chromatin in Archaea. », Science, vol. 357, no 6351, , p. 609-612 (PMID 28798133, PMCID PMC5747315, DOI 10.1126/science.aaj1849)
- (en) Bram Henneman, Clara van Emmerik, Hugo van Ingen et Remus T. Dame, « Structure and function of archaeal histones. », PLoS Genetics, vol. 14, no 9, , e1007582 (PMID 30212449, PMCID PMC6136690, DOI 10.1371/journal.pgen.1007582)
- (en) Richard Ward, Andrew Bowman, Hassane El-Mkami, Tom Owen-Hughes et David G. Norman, « Long distance PELDOR measurements on the histone core particle. », Journal of the American Chemical Society, vol. 131, no 4, , p. 1348-9 (PMID 19138067, PMCID PMC3501648, DOI 10.1021/ja807918f)
- (en) Erica L. Mersfelder et Mark R. Parthun, « The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. », Nucleic Acids Research, vol. 34, no 9, , p. 2653-62 (PMID 16714444, PMCID PMC1464108, DOI 10.1093/nar/gkl338)
- (en) Philipp Tropberger et Robert Schneider, « Scratching the (lateral) surface of chromatin regulation by histone modifications. », Nature Structural & Molecular Biology, vol. 20, no 6, , p. 657-61 (PMID 23739170, DOI 10.1038/nsmb.2581)
- (en) Lizabeth A. Allison, Fundamental Molecular Biology, Hoboken, New Jersey, États-Unis, John Wiley & Sons, , 2e éd., XXVI-656 p. (ISBN 978-1-118-05981-4)
- (en) Robert Klose et Yi Zhang, « Regulation of histone methylation by demethylimination and demethylation », Nature Reviews. Molecular and Cell Biology, vol. 8, no 4, , p. 307-18 (PMID 17342184, DOI 10.1038/nrm2143)
- (en) Thomas Jenuwein et C. David Allis, « Translating the histone code. », Science, vol. 293, no 5532, , p. 1074-80 (PMID 11498575, DOI 10.1126/science.1063127)
- (en) Ning Song, Jie Liu, Shucai An, Tomoya Nishino, Yoshitaka Hishikawa et Takehiko Koji, « Immunohistochemical analysis of histone H3 modifications in germ cells during mouse spermatogenesis. », Acta Histochemica et Cytochemica, vol. 44, no 4, , p. 183-90 (PMID 21927517, PMCID PMC3168764, DOI 10.1267/ahc.11027)
- (en) Elizaveta V. Benevolenskaya, « Histone H3K4 demethylases are essential in development and differentiation. », Biochemistry and Cell Biology, vol. 85, no 4, , p. 435-43 (PMID 17713579, DOI 10.1139/O07-057)
- (en) Artem Barski, Suresh Cuddapah, Kairong Cui, Tae-Young Roh, Dustin E. Schones, Zhibin Wang, Gang Wei, Iouri Chepelev et Keji Zhao, « High-resolution profiling of histone methylations in the human genome. », Cell, vol. 129, no 4, , p. 823-37 (PMID 17512414, DOI 10.1016/j.cell.2007.05.009)
- (en) David J. Steger, Martina I. Lefterova, Lei Ying, Aaron J. Stonestrom, Michael Schupp, David Zhuo, Adam L. Vakoc, Ja-Eun Kim, Junjie Chen, Mitchell A. Lazar, Gerd A. Blobel et Christopher R. Vakoc, « DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. », Molecular and Cellular Biology, vol. 28, no 8, , p. 2825-39 (PMID 18285465, PMCID PMC2293113, DOI 10.1128/MCB.02076-07)
- (en) Jeffrey A. Rosenfeld, Zhibin Wang, Dustin E. Schones, Keji Zhao, Rob DeSalle et Michael Q. Zhang, « Determination of enriched histone modifications in non-genic portions of the human genome. », BMC Genomics, vol. 10, , p. 143 (PMID 19335899, PMCID PMC2667539, DOI 10.1186/1471-2164-10-143)
- (en) Christoph M. Koch, Robert M. Andrews, Paul Flicek, Shane C. Dillon, Ulas Karaoz, Gayle K. Clelland, Sarah Wilcox, David M. Beare, Joanna C. Fowler, Phillippe Couttet, Keith D. James, Gregory C. Lefebvre, Alexander W. Bruce, Oliver M. Dovey, Peter D. Ellis, Pawandeep Dhami, Cordelia F. Langford, Zhiping Weng, Ewan Birney, Nigel P. Carter, David Vetrie et Ian Dunham, « The landscape of histone modifications across 1% of the human genome in five human cell lines. », Genome Research, vol. 17, no 6, , p. 691-707 (PMID 17567990, PMCID PMC1891331, DOI 10.1101/gr.5704207)
- (en) Benoit Guillemette, Paul Drogaris, Hsiu-Hsu Sophia Lin, Harry Armstrong, Kyoko Hiragami-Hamada, Axel Imhof, Eric Bonneil, Pierre Thibault, Alain Verreault et Richard J. Festenstein, « H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. », PLoS Genetics, vol. 7, no 3, , e1001354 (PMID 21483810, PMCID PMC3069113, DOI 10.1371/journal.pgen.1001354)
- (en) Menno P. Creyghton, Albert W. Cheng, G. Grant Welstead, Tristan Kooistra, Bryce W. Carey, Eveline J. Steine, Jacob Hanna, Michael A. Lodato, Garrett M. Frampton, Phillip A. Sharp, Laurie A. Boyer, Richard A. Young et Rudolf Jaenisch, « Histone H3K27ac separates active from poised enhancers and predicts developmental state. », Proceedings of the National Academy of Sciences of the United States of America, vol. 107, no 50, , p. 21931-6 (PMID 21106759, PMCID PMC3003124, DOI 10.1073/pnas.1016071107)
- (en) Lorna A. Farrelly, Robert E. Thompson, Shuai Zhao, Ashley E. Lepack, Yang Lyu, Natarajan V. Bhanu, Baichao Zhang, Yong-Hwee E. Loh, Aarthi Ramakrishnan, Krishna C. Vadodaria, Kelly J. Heard, Galina Erikson, Tomoyoshi Nakadai, Ryan M Bastle, Bradley J. Lukasak, Henry 3rd Zebroski, Natalia Alenina, Michael Bader, Olivier Berton, Robert G. Roeder, Henrik Molina, Fred H. Gage, Li Shen, Benjamin A. Garcia, Haitao Li, Tom W. Muir et Ian Maze, « Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. », Nature, vol. 567, no 7749, , p. 535-539 (PMID 30867594, PMCID PMC6557285, DOI 10.1038/s41586-019-1024-7)
- (en) Maria A. Christophorou, Goncalo Castelo-Branco, Richard P. Halley-Stott, Clara Slade Oliveira, Remco Loos, Aliaksandra Radzisheuskaya, Kerri A. Mowen, Paul Bertone, Jose C. R. Silva, Magdalena Zernicka-Goetz, Michael L. Nielsen, John B. Gurdon et Tony Kouzarides, « Citrullination regulates pluripotency and histone H1 binding to chromatin. », Nature, vol. 507, no 7490, , p. 104-8 (PMID 24463520, PMCID PMC4843970, DOI 10.1038/nature12942)
- (en) Graeme L. Cuthbert, Sylvain Daujat, Andrew W. Snowden, Hediye Erdjument-Bromage, Teruki Hagiwara, Michiyuki Yamada, Robert Schneider, Philip D. Gregory, Paul Tempst, Andrew J. Bannister et Tony Kouzarides, « Histone deimination antagonizes arginine methylation. », Cell, vol. 118, no 5, , p. 545-53 (PMID 15339660, DOI 10.1016/j.cell.2004.08.020)
- (en) Nevan J. Krogan, Jim Dover, Adam Wood, Jessica Schneider, Jonathan Heidt, Marry Ann Boateng, Kimberly Dean, Owen W. Ryan, Ashkan Golshani, Mark Johnston, Jack F. Greenblatt et Ali Shilatifard, « The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. », Molecular Cell, vol. 11, no 3, , p. 721-9 (PMID 12667454, DOI 10.1016/s1097-2765(03)00091-1)
- (en) Huck Hui Ng, François Robert, Richard A. Young et Kevin Struhl, « Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. », Molecular Cell, vol. 11, no 3, , p. 709-19 (PMID 12667453, DOI 10.1016/s1097-2765(03)00092-3)
- (en) Bradley E. Bernstein, Michael Kamal, Kerstin Lindblad-Toh, Stefan Bekiranov, Dione K. Bailey, Dana J. Huebert, Scott McMahon, Elinor K. Karlsson, Edward J. 3rd Kulbokas, Thomas R. Gingeras, Stuart L. Schreiber et Eric S. Lander, « Genomic maps and comparative analysis of histone modifications in human and mouse. », Cell, vol. 120, no 2, , p. 169-81 (PMID 15680324, DOI 10.1016/j.cell.2005.01.001)
- (en) Nevan J. Krogan, Jim Dover, Shahram Khorrami, Jack F. Greenblatt, Jessica Schneider, Mark Johnston et Ali Shilatifard, « COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. », Journal of Biological Chemistry, vol. 277, no 13, , p. 10753-5 (PMID 11805083, DOI 10.1074/jbc.C200023200)
- (en) Assen Roguev, Daniel Schaft, Anna Shevchenko, W. W. M. Pim Pijnappel, Matthias Wilm, Rein Aasland et A. Francis Stewart, « The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. », EMBO Journal, vol. 20, no 24, , p. 7137-48 (PMID 11742990, PMCID PMC125774, DOI 10.1093/emboj/20.24.7137)
- (en) Peter L. Nagy, Joachim Griesenbeck, Roger D. Kornberg et Michael L. Cleary, « A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. », Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no 1, , p. 90-4 (PMID 11752412, PMCID PMC117519, DOI 10.1073/pnas.221596698)
- (en) Adam Wood, Jessica Schneider, Jim Dover, Mark Johnston et Ali Shilatifard, « The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. », Molecular Cell, vol. 20, no 4, , p. 589-99 (PMID 16307922, DOI 10.1016/j.molcel.2005.09.010)
- (en) Boris Sarcevic, Amanda Mawson, Rohan T. Baker et Robert L. Sutherland, « Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. », EMBO Journal, vol. 21, no 8, , p. 2009-18 (PMID 11953320, PMCID PMC125963, DOI 10.1093/emboj/21.8.2009)
- (en) Kenneth Robzyk, Judith Recht et Mary Ann Osley, « Rad6-dependent ubiquitination of histone H2B in yeast. », Science, vol. 287, no 5452, , p. 501-4 (PMID 10642555, DOI 10.1126/science.287.5452.501)
- (en) Zu-Wen Sun et C. David Allis, « Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. », Nature, vol. 418, no 6893, , p. 104-8 (PMID 12077605, DOI 10.1038/nature00883)
- (en) Jim Dover, Jessica Schneider, Mary Anne Tawiah-Boateng, Adam Wood, Kimberly Dean, Mark Johnston et Ali Shilatifard, « Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. », Journal of Biological Chemistry, vol. 277, no 32, , p. 28368-71 (PMID 12070136, DOI 10.1074/jbc.C200348200)
- (en) Brian D. Strahl, Patrick A. Grant, Scott D. Briggs, Zu-Wen Sun, James R. Bone, Jennifer A. Caldwell, Sahana Mollah, Richard G. Cook, Jeffrey Shabanowitz, Donald F. Hunt et C. David Allis, « Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. », Molecular and Cellular Biology, vol. 22, no 5, , p. 1298-306 (PMID 11839797, PMCID PMC134702, DOI 10.1128/mcb.22.5.1298-1306.2002)
- (en) Jiaxu Li, Danesh Moazed et Steven P. Gygi, « Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. », Journal of Biological Chemistry, vol. 277, no 51, , p. 49383-8 (PMID 12381723, DOI 10.1074/jbc.M209294200)
- (en) Michael J. Carrozza, Bing Li, Laurence Florens, Tamaki Suganuma, Selene K. Swanson, Kenneth K. Lee, Wei-Jong Shia, Scott Anderson, John Yates, Michael P. Washburn et Jerry L. Workman, « Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. », Cell, vol. 123, no 4, , p. 581-92 (PMID 16286007, DOI 10.1016/j.cell.2005.10.023)
- (en) Michael-Christopher Keogh, Siavash K. Kurdistani, Stephanie A. Morris, Seong Hoon Ahn, Vladimir Podolny, Sean R. Collins, Maya Schuldiner, Kayu Chin, Thanuja Punna, Natalie J. Thompson, Charles Boone, Andrew Emili, Jonathan S. Weissman, Timothy R. Hughes, Brian D. Strahl, Michael Grunstein, Jack F. Greenblatt, Stephen Buratowski et Nevan J. Krogan, « Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. », Cell, vol. 123, no 4, , p. 593-605 (PMID 16286008, DOI 10.1016/j.cell.2005.10.025)
- (en) Amita A. Joshi et Kevin Struhl, « Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. », Molecular Cell, vol. 20, no 6, , p. 971-8 (PMID 16364921, DOI 10.1016/j.molcel.2005.11.021)
- (en) Andrei Kuzmichev, Kenichi Nishioka, Hediye Erdjument-Bromage, Paul Tempst et Danny Reinberg, « Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. », Genes & Development, vol. 16, no 22, , p. 2893-905 (PMID 12435631, PMCID PMC187479, DOI 10.1101/gad.1035902)
- (en) Ru Cao, Liangjun Wang, Hengbin Wang, Li Xia, Hediye Erdjument-Bromage, Paul Tempst, Richard S. Jones et Yi Zhang, « Role of histone H3 lysine 27 methylation in Polycomb-group silencing. », Science, vol. 298, no 5595, , p. 1039-43 (PMID 12351676, DOI 10.1126/science.1076997)
- (en) Mariana de Napoles, Jacqueline E. Mermoud, Rika Wakao, Y. Amy Tang, Mitusuhiro Endoh, Ruth Appanah, Tatyana B. Nesterova, Jose Silva, Arie P. Otte, Miguel Vidal, Haruhiko Koseki et Neil Brockdorff, « Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. », Developmental Cell, vol. 7, no 5, , p. 663-76 (PMID 15525528, DOI 10.1016/j.devcel.2004.10.005)
- (en) Hengbin Wang, Liangjun Wang, Hediye Erdjument-Bromage, Miguel Vidal, Paul Tempst, Richard S. Jones et Yi Zhang, « Role of histone H2A ubiquitination in Polycomb silencing. », Nature, vol. 431, no 7010, , p. 873-8 (PMID 15386022, DOI 10.1038/nature02985)
- (en) Ligia Tavares, Emilia Dimitrova, David Oxley, Judith Webster, Raymond Poot, Jeroen Demmers, Karel Bezstarosti, Stephen Taylor, Hiroki Ura, Hiroshi Koide, Anton Wutz, Miguel Vidal, Sarah Elderkin et Neil Brockdorff, « RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. », Cell, vol. 148, no 4, , p. 664-78 (PMID 22325148, PMCID PMC3281992, DOI 10.1016/j.cell.2011.12.029)
- (en) Zhonghua Gao, Jin Zhang, Roberto Bonasio, Francesco Strino, Ayana Sawai, Fabio Parisi, Yuval Kluger et Danny Reinberg, « PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. », Molecular Cell, vol. 45, no 3, , p. 344-56 (PMID 22325352, PMCID PMC3293217, DOI 10.1016/j.molcel.2012.01.002)
- (en) Andre Verdel, Songtao Jia, Scott Gerber, Tomoyasu Sugiyama, Steven Gygi, Shiv I. S. Grewal et Danesh Moazed, « RNAi-mediated targeting of heterochromatin by the RITS complex. », Science, vol. 303, no 5658, , p. 672-6 (PMID 14704433, PMCID PMC3244756, DOI 10.1126/science.1093686)
- (en) Stephen Rea, Frank Eisenhaber, Dónal O’Carroll, Brian D. Strahl, Zu-Wen Sun, Manfred Schmid, Susanne Opravil, Karl Mechtler, Chris P. Ponting, C. David Allis et Thomas Jenuwein, « Regulation of chromatin structure by site-specific histone H3 methyltransferases. », Nature, vol. 406, no 6796, , p. 593-9 (PMID 10949293, DOI 10.1038/35020506)
- (en) Andreq J. Bannister, Philip Zegerman, Janet F. Partridge, Eric A. Miska, Jean O. Thomas, Robin C. Allshire et Tony Kouzarides, « Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. », Nature, vol. 410, no 6824, , p. 120-4 (PMID 11242054, DOI 10.1038/35065138)
- (en) Monika Lachner, Dónal O’Carroll, Stephen Rea, Karl Mechtler et Thomas Jenuwein, « Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. », Nature, vol. 410, no 6824, , p. 116-20 (PMID 11242053, DOI 10.1038/35065132)
- (en) Gaurav Bajpai, Ishutesh Jain, Mandar M. Inamdar, Dibyendu Das et Ranjith Padinhateeri, « Binding of DNA-bending non-histone proteins destabilizes regular 30-nm chromatin structure. », PLoS Computational Biology, vol. 13, no 1, , e1005365 (PMID 28135276, PMCID PMC5305278, DOI 10.1371/journal.pcbi.1005365)
- (en) Gunnar Schotta, Monika Lachner, Kavitha Sarma, Anja Ebert, Roopsha Sengupta, Gunter Reuter, Danny Reinberg et Thomas Jenuwein, « A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. », Genes & Development, vol. 18, no 11, , p. 1251-62 (PMID 15145825, PMCID PMC420351, DOI 10.1101/gad.300704)
- (en) Niki Kourmouli, Peter Jeppesen, Shantha Mahadevhaiah, Paul Burgoyne, Rong Wu, David M. Gilbert, Silvia Bongiorni, Giorgio Prantera, Laura Fanti, Sergio Pimpinelli, Wei Shi, Reinald Fundele et Prim B. Singh, « Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. », Journal of Cell Science, vol. 117, no Pt 12, , p. 2491-501 (PMID 15128874, DOI 10.1242/jcs.01238)
- (en) Bradley E. Bernstein, Tarjei S. Mikkelsen, Xiaohui Xie, Michael Kamal, Dana J. Huebert, James Cuff, Ben Fry, Alex Meissner, Marius Wernig, Kathrin Plath, Rudolf Jaenisch, Alexandre Wagschal, Robert Feil, Stuart L. Schreiber et Eric S Lander, « A bivalent chromatin structure marks key developmental genes in embryonic stem cells. », Cell, vol. 125, no 2, , p. 315-26 (PMID 16630819, DOI 10.1016/j.cell.2006.02.041)
- (en) Emmy P. Rogakou, Duane R. Pilch, Ann H. Orr, Vessela S. Ivanova et William M. Bonner, « DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. », Journal of Biological Chemistry, vol. 273, no 10, , p. 5858-68 (PMID 9488723, DOI 10.1074/jbc.273.10.5858)
- (en) Tanya T. Paull, Emmy P. Rogakou, Vikky Yamazaki, Cordula U. Kirchgessner, Martin Gellert et William M. Bonner, « A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. », Current Biology, vol. 10, no 15, , p. 886-95 (PMID 10959836, DOI 10.1016/s0960-9822(00)00610-2)
- (en) Arkady Celeste, Simone Petersen, Peter J. Romanienko, Oscar Fernandez-Capetillo, Hua Tang Chen, Olga A. Sedelnikova, Bernardo Reina-San-Martin, Vincenzo Coppola, Eric Meffre, Michael J. Difilippantonio, Christophe Redon, Duane R. Pilch, Alexandru Olaru, Michael Eckhaus, R. Daniel Camerini-Otero, Lino Tessarollo, Ferenc Livak, Katia Manova, William M. Bonner, Michel C. Nussenzweig et Andre Nussenzweig, « Genomic instability in mice lacking histone H2AX. », Science, vol. 296, no 5569, , p. 922-7 (PMID 11934988, PMCID PMC4721576, DOI 10.1126/science.1069398)
- (en) Robert Shroff, Ayelet Arbel-Eden, Duane Pilch, Grzegorz Ira, William M. Bonner, John H. Petrini, James E. Haber et Michael Lichten, « Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. », Current Biology, vol. 14, no 19, , p. 1703-11 (PMID 15458641, PMCID PMC4493763, DOI 10.1016/j.cub.2004.09.047)
- (en) Emmy P. Rogakou, Chye Boon, Christophe Redon et William M. Bonner, « Megabase chromatin domains involved in DNA double-strand breaks in vivo. », Journal of Cell Biology, vol. 146, no 5, , p. 905-16 (PMID 10477747, PMCID PMC2169482, DOI 10.1083/jcb.146.5.905)
- (en) Grant S. Stewart, Bin Wang, Colin R. Bignell, A Malcolm R. Taylor et Stephen J. Elledge, « MDC1 is a mediator of the mammalian DNA damage checkpoint. », Nature, vol. 421, no 6926, , p. 961-6 (PMID 12607005, DOI 10.1038/nature01446)
- (en) Simon Bekker-Jensen et Niels Mailand, « Assembly and function of DNA double-strand break repair foci in mammalian cells. », DNA Repair, vol. 9, no 12, , p. 1219-28 (PMID 21035408, DOI 10.1016/j.dnarep.2010.09.010)
- (en) Anil Ozdemir, Salvatore Spicuglia, Edwin Lasonder, Michiel Vermeulen, Coen Campsteijn, Hendrik G. Stunnenberg et Colin Logie, « Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. », Journal of Biological Chemistry, vol. 280, no 28, , p. 25949-52 (PMID 15888442, DOI 10.1074/jbc.C500181200)
- (en) Hiroshi Masumoto, David Hawke, Ryuji Kobayashi et Alain Verreault, « A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. », Nature, vol. 436, no 7048, , p. 294-8 (PMID 16015338, DOI 10.1038/nature03714)
- (en) Robert Driscoll, Amanda Hudson et Stephen P. Jackson, « Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. », Science, vol. 315, no 5812, , p. 649-52 (PMID 17272722, PMCID PMC3334813, DOI 10.1126/science.1135862)
- (en) Junhong Han, Hui Zhou, Bruce Horazdovsky, Kangling Zhang, Rui-Ming Xu et Zhiguo Zhang, « Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. », Science, vol. 315, no 5812, , p. 653-5 (PMID 17272723, DOI 10.1126/science.1133234)
- (en) Chandrima Das, M. Scott Lucia, Kirk C. Hansen et Jessica K. Tyler, « CBP/p300-mediated acetylation of histone H3 on lysine 56. », Nature, vol. 459, no 7243, , p. 113-7 (PMID 19270680, PMCID PMC2756583, DOI 10.1038/nature07861)
- (en) Junhong Han, Hui Zhou, Zhizhong Li, Rui-Ming Xu et Zhiguo Zhang, « Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. », Journal of Biological Chemistry, vol. 282, no 39, , p. 28587-96 (PMID 17690098, DOI 10.1074/jbc.M702496200)
- (en) Hugo Wurtele, Gitte Schalck Kaiser, Julien Bacal, Edlie St-Hilaire, Eun-Hye Lee, Sarah Tsao, Jonas Dorn, Paul Maddox, Michael Lisby, Philippe Pasero et Alain Verreault, « Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. », Molecular and Cellular Biology, vol. 32, no 1, , p. 154-72 (PMID 22025679, PMCID PMC3255698, DOI 10.1128/MCB.05415-11)
- (en) Hugo Wurtele, Sarah Tsao, Guylaine Lepine, Alaka Mullick, Jessy Tremblay, Paul Drogaris, Eun-Hye Lee, Pierre Thibault, Alain Verreault et Martine Raymond, « Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. », Nature Medicine, vol. 16, no 7, , p. 774-80 (PMID 20601951, PMCID PMC4108442, DOI 10.1038/nm.2175)
- (en) Feng Li, Guogen Mao, Dan Tong, Jian Huang, Liya Gu, Wei Yang et Guo-Min Li, « The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. », Cell, vol. 153, no 3, , p. 590-600 (PMID 23622243, PMCID PMC3641580, DOI 10.1016/j.cell.2013.03.025)
- (en) Fran Supek et Ben Lehner, « Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. », Cell, vol. 170, no 3, , p. 534-547.e23 (PMID 28753428, DOI 10.1016/j.cell.2017.07.003)
- (en) Bryan J. Wilkins, Nils A. Rall, Yogesh Ostwal, Tom Kruitwagen, Kyoko Hiragami-Hamada, Marco Winkler, Yves Barral, Wolfgang Fischle et Heinz Neumann, « A cascade of histone modifications induces chromatin condensation in mitosis. », Science, vol. 343, no 6166, , p. 77-80 (PMID 24385627, DOI 10.1126/science.1244508)
- (en) Kristen M. Johansen et Jorgen Johansen, « Regulation of chromatin structure by histone H3S10 phosphorylation. », Chromosome Research, vol. 14, no 4, , p. 393-404 (PMID 16821135, DOI 10.1007/s10577-006-1063-4)
- (en) Maikel Castellano-Pozo, Jose M. Santos-Pereira, Ana G. Rondon, Sonia Barroso, Eloisa Andujar, Monica Perez-Alegre, Tatiana Garcia-Muse et Andres Aguilera, « R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. », Molecular Cell, vol. 52, no 4, , p. 583-90 (PMID 24211264, DOI 10.1016/j.molcel.2013.10.006)
- (en) Wang L. Cheung, Kozo Ajiro, Kumiko Samejima, Malgorzata Kloc, Peter Cheung, Craig A. Mizzen, Alexander Beeser, Laurence D. Etkin, Jonathan Chernoff, William C. Earnshaw et C. David Allis, « Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. », Cell, vol. 113, no 4, , p. 507-17 (PMID 12757711, DOI 10.1016/s0092-8674(03)00355-6)
- (en) Sung-Hee Ahn, Wang L. Cheung, Jer-Yuan Hsu, Robert L. Diaz, M Mitchell Smith et C David Allis, « Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. », Cell, vol. 120, no 1, , p. 25-36 (PMID 15652479, DOI 10.1016/j.cell.2004.11.016)
- (en) Alfred J. Robison et Eric J. Nestler, « Transcriptional and epigenetic mechanisms of addiction. », Nature Reviews. Neuroscience, vol. 12, no 11, , p. 623-37 (PMID 21989194, PMCID PMC3272277, DOI 10.1038/nrn3111)
- (en) Leah N. Hitchcock et K Matthew Lattal, « Histone-mediated epigenetics in addiction. », Progress in Molecular Biology and Translational Science, vol. 128, , p. 51-87 (PMID 25410541, PMCID PMC5914502, DOI 10.1016/B978-0-12-800977-2.00003-6)
- (en) Susan C. McQuown et Marcelo A. Wood, « Epigenetic regulation in substance use disorders. », Current Psychiatry Reports, vol. 12, no 2, , p. 145-53 (PMID 20425300, PMCID PMC2847696, DOI 10.1007/s11920-010-0099-5)
- (en) National Institute on Drug Abuse, « Is nicotine addictive? », sur drugabuse.gov, (consulté le )
- (en) Amir Levine, Yanyou Huang, Bettina Drisaldi, Edmund A. Griffin Jr., Daniela D. Pollak, Shiqin Xu, Deqi Yin, Christine Schaffran, Denise B. Kandel et Eric R. Kandel, « Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. », Science Translational Medicine, vol. 3, no 107, , p. 107ra109 (PMID 22049069, PMCID PMC4042673, DOI 10.1126/scitranslmed.3003062)
- (en) James K. Ruffle, « Molecular neurobiology of addiction: what's all the (Delta)FosB about? », American Journal of Drug and Alcohol Abuse, vol. 40, no 6, , p. 428-37 (PMID 25083822, DOI 10.3109/00952990.2014.933840)
- (en) Eric J. Nestler, Michel Barrot et David W. Self, « ΔFosB: a sustained molecular switch for addiction. », Proceedings of the National Academy of Sciences of the United States of America, vol. 98, no 20, , p. 11042-6 (PMID 11572966, PMCID PMC58680, DOI 10.1073/pnas.191352698)
- (en) Claudio D’Addario, Francesca F. Caputi, Tomas J. Ekstrom, Manuela Di Benedetto, Mauro Maccarrone, Patrizia Romualdi et Sanzio Candeletti, « Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. », Journal of Molecular Neuroscience, vol. 49, no 2, , p. 312-9 (PMID 22684622, DOI 10.1007/s12031-012-9829-y)
- (en) National Institute on Drug Abuse, « What is the scope of methamphetamine abuse in the United States? », sur drugabuse.gov, (consulté le )
- (en) Arthur Godino, Subramaniam Jayanthi et Jean Lud Cadet, « Epigenetic landscape of amphetamine and methamphetamine addiction in rodents. », Epigenetics, vol. 10, no 7, , p. 574-80 (PMID 26023847, PMCID PMC4622560, DOI 10.1080/15592294.2015.1055441)
- (en) Fabio C. Cruz, F. Javier Rubio et Bruce T. Hope, « Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. », Brain Research, vol. 1628, no Pt A, , p. 157-73 (PMID 25446457, PMCID PMC4427550, DOI 10.1016/j.brainres.2014.11.005)
- (en) Joseph Hannon Bozorgmehr, « The origin of chromosomal histones in a 30S ribosomal protein. », Gene, vol. 726, , p. 144155 (PMID 31629821, DOI 10.1016/j.gene.2019.144155)
- (en) Peter J. Rizzo, « Those amazing dinoflagellate chromosomes. », Cell Research, vol. 13, no 4, , p. 215-7 (PMID 12974611, DOI 10.1038/sj.cr.7290166)
- (en) Paul B. Talbert et Steven Henikoff, « Chromatin: packaging without nucleosomes. », Current Biology, vol. 22, no 24, , R1040-3 (PMID 23257187, DOI 10.1016/j.cub.2012.10.052)
- (en) Harold E. Kasinsky, John D. Lewis, Joel B. Dacks et Juan Ausió, « Origin of H1 linker histones. », FASEB Journal, vol. 15, no 1, , p. 34-42 (PMID 11149891, DOI 10.1096/fj.00-0237rev)
- (en) Hugh J. Clarke, « Nuclear and chromatin composition of mammalian gametes and early embryos. », Biochemistry and Cell Biology, vol. 70, nos 10-11, , p. 856-66 (PMID 1297351, DOI 10.1139/o92-134)
- (en) Benoit Guillemette, Alain R. Bataille, Nicolas Gevry, Maryse Adam, Mathieu Blanchette, François Robert et Luc Gaudreau, « Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. », PLoS Biology, vol. 3, no 12, , e384 (PMID 16248679, PMCID PMC1275524, DOI 10.1371/journal.pbio.0030384)
- (en) Kami Ahmad et Steven Henikoff, « The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. », Molecular Cell, vol. 9, no 6, , p. 1191-200 (PMID 12086617, DOI 10.1016/s1097-2765(02)00542-7)
- (en) Robertus A. M. de Bruin, W. Hayes McDonald, Tatyana I. Kalashnikova, John 3rd Yates et Curt Wittenberg, « Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. », Cell, vol. 117, no 7, , p. 887-98 (PMID 15210110, DOI 10.1016/j.cell.2004.05.025)
- (en) Haixin Xu, Ung-Jin Kim, Tillman Schuster et Michael Grunstein, « Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. », Molecular and Cellular Biology, vol. 12, no 11, , p. 5249-59 (PMID 1406694, PMCID PMC360458, DOI 10.1128/mcb.12.11.5249)
- (en) Dessislava. Dimova, Zeena Nackerdien, Seth Furgeson, Sayaka Eguchi et Mary Ann Osley, « A role for transcriptional repressors in targeting the yeast Swi/Snf complex. », Molecular Cell, vol. 4, no 1, , p. 75-83 (PMID 10445029, DOI 10.1016/s1097-2765(00)80189-6)
- (en) Zbigniew Dominski, Judith A. Erkmann, Xiaocui Yang, Ricardo Sanchez et William F. Marzluff, « A novel zinc finger protein is associated with U7 snRNP and interacts with the stem-loop binding protein in the histone pre-mRNP to stimulate 3'-end processing. », Genes & Development, vol. 16, no 1, , p. 58-71 (PMID 11782445, PMCID PMC155312, DOI 10.1101/gad.932302)
- (en) Zbigniew Dominski, Xiao-cui Yang, Handan Kaygun, Michal Dadlez et William F. Marzluff, « A 3' exonuclease that specifically interacts with the 3' end of histone mRNA. », Molecular Cell, vol. 12, no 2, , p. 295-305 (PMID 14536070, DOI 10.1016/s1097-2765(03)00278-8)
- (en) Lianxing Zheng, Zbigniew Dominski, Xiao-Cui Yang, Phillip Elms, Christy S. Raska, Christoph H. Borchers et William F. Marzluff, « Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. », Molecular and Cellular Biology, vol. 23, no 5, , p. 1590-601 (PMID 12588979, PMCID PMC151715, DOI 10.1128/mcb.23.5.1590-1601.2003)
- (en) Qiuyan Wang, Iain A. Sawyer, Myong-Hee Sung, David Sturgill, Sergey P. Shevtsov, Gianluca Pegoraro, Ofir Hakim, Songjoon Baek, Gordon L. Hager et Miroslav Dundr, « Cajal bodies are linked to genome conformation. », Nature Communications, vol. 7, , p. 10966 (PMID 26997247, PMCID PMC4802181, DOI 10.1038/ncomms10966)
- (en) Jiyong Zhao, Brian K. Kennedy, Brandon D. Lawrence, David A. Barbie, A. Gregory Matera, Jonathan A. Fletcher et Ed Harlow, « NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. », Genes & Development, vol. 14, no 18, , p. 2283-97 (PMID 10995386, PMCID PMC316937, DOI 10.1101/gad.827700)
